This is the 3rd in a three-part series from Medscape on the effect of AI on drug discovery and advancement. Part 1 is about AI’s function in developing faster, more reliable medical trials. Part 2 is about the usage of AI to discover brand-new applications for existing drugs.
What if we informed you that the verysame innovation behind deepfake pictures might quickly bring brand-new and enhanced drugs to your clients?
Unlike some of those pictures, that’s not phony news.
Researchers are loaning from synthetic intelligence (AI) designs, such as OpenAI’s DALL-E, that equate text into images to style never-before-seen proteins from scratch. These styles might kind the basis of future drugs or vaccines.

“For the veryfirst time, we are seeing AI systems able to create extremely reasonable protein structures in a manageable method in seconds,” stated Namrata Anand, PhD, a bioengineer and computersystem researcher.
In 2022, Anand utilized this innovation to establish the design that generated her business Diffuse Bio. Also that year, a de novo protein developed with AI by scientists at the University of Washington, Seattle, acquired scientific approval abroad as a COVID-19 vaccine. Biotech business Absci’s de novo antibody for inflammatory bowel illness is goingthrough investigational brand-new drug-enabling researchstudies in hopes of beginning medical trials in2025 Other AI-enhanced efforts focus on developing understood proteins or finding methods to target proteins thoughtabout undruggable.
The innovation is early, however the tipping point is here. These advances are start to have concrete results on drug discovery and advancement.
CEO Sean McClain anticipates Absci’s AI-enhanced technique to slash the time and expenses required to bring brand-new particles to the center, from a 5-and-a-half-year procedure costing up to $100 million to a 2-year procedure costing $15 million.
“We’re actually beginning to break the biotech economics,” McClain stated. “Instead of investing typically $100 million into one drug property, you can now invest it into 5 to 10 and get them into the center much quicker.”
Starting From Scratch
Proteins— there are about 20,000 in the human body — dealwith much of the heavy lifting needed for life. They offer cells structure however likewise sendout essential messages and procedure signals. Some proteins help embryonic advancement or stop cells from turning malignant. Disease-fighting antibodies are proteins. So are enzymes and lotsof hormonalagents.

Proteins are formed by strings of electrically charged amino acids that bringin or wardoff one another, triggering folding that provides proteins unique shapes. These shapes aid identify each protein’s function.
Researchers have long lookedfor to comprehend how proteins fold into shape in hopes of knowing how to construct them from scratch. Even as justrecently as 2017 — the year of a flu-fighting protein style development — protein style was still relatively ineffective.
“The softwareapplication simply would not work dependably every single time that you’d desire it to,” Anand stated.
But AI has because taken off. In 2022, the open-access AlphaFold Protein Structure Database, established by Google DeepMind and the European Bioinformatics Institute, made a chest of more than 200 million protein structure forecasts searchable for any scientist. In drug discovery, these protein structures can supply beginning points for finding an preliminary “hit” to enhance.
This “high-quality, big dataset of proteins” is the just one of its kind, stated Gregory Bowman, PhD, a teacher at the University of Pennsylvania, in Philadelphia. “But it doesn’t mean take AlphaFold, change brand-new drug discovery, and presto, get a lot of drugs.”
A capacity snag: “A big portion of proteins are thoughtabout undruggable,” Bowman stated. That is, they absence cavities where drugs might bind or those cavities are too subtle for most innovations to discover.
Bowman’s laboratory utilized computersystem simulations to hunt down those “cryptic pockets” and then followed up with experiments to verify their forecasts. They constructed a dataset and utilized it to train a neural network, called PocketMiner, which utilizes device knowing to anticipate where puzzling pockets may lie.

Last year, PocketMiner anticipated puzzling pocket areas in almost 3 lots cancer-related protein structures, opening the door for capacity brand-new treatments.
“I think these puzzling pockets are in most proteins, which implies that they’re appropriate to most cancers,” Bowman stated.
His laboratory is teamingup with start-up business on breast cancer and uveal cancermalignancy, a cancer of the eye. They’re likewise checkingout targets for illness such as leukemia and pancreatic cancer.
That researchstudy can enhance de novo style efforts, stated Anand. “In my mind, they’re 2 cutsinhalf of the verysame coin. If you have extremely great forecast of where to bind, then you can style a drug versus stated pocket.”
And if scientists produce thousands of de novo proteins, a strong predictive design can aid identify which style is more mostlikely to work. “There’s a method to weave them together,” Anand stated.
Generating Protein Structures and Sequences
Some proteins are more difficult to style than others. Take helical peptides — brief, versatile, coil-like chains of amino acids discovered in really little concentrations in the body. Designing proteins that can bind to such evasive structures hasactually been a battle. But a group at Baker Lab, part of the Institute for Protein Design at the University of Washington, utilized 2 deep knowing tools to pull it off.
Their efforts, released last December, might make identifying illness lessexpensive and muchfaster, and lead to therapies that bind to hormonalagents to block their activity. For circumstances, their developed proteins target parathyroid hormonalagent (PTH), a parathyroid cancer biomarker, and neuropeptide Y, associated with Alzheimer’s illness. Their de novo protein found PTH by mass spectrometry and allowed protein biosensors responsive to PTH.
“Making antibodies for some of these targets, which are the gold-standard particles for diagnostics or rehabs, might take months,” stated lead author Susana Vazquez Torres, PhD, a scientist at the Baker Lab. But with AI, scientists can produce thousands of proteins in simply one day.

They start with a target helical peptide (such as PTH) surrounded by a random circulation of atoms, like “a cloud of sound.” Next, a diffusion design “de-noises” the atoms, triggering them to fold into the shape of a protein. Again, this innovation obtains from the AI-powered text-to-image designs pointedout earlier, which are “trained” on troves of online images and text.
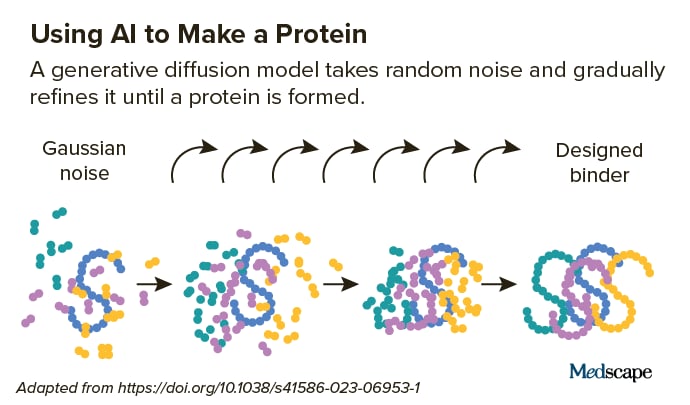
“Diffusion is a really effective innovation since you wear’t necessaril